EDEN: 28 Billion Parameters for Programming Biology
Bringing a new drug to market takes roughly 10 to 15 years and costs an estimated $2.5 billion 1. Most candidates fail. The ones that succeed often trace back to a molecule someone found in nature, tweaked in a lab, and pushed through a decade of trials. The bottleneck isn't ambition or funding. It's that therapeutic design remains stubbornly artisanal: identify a target, screen millions of candidates against it, optimize a handful in the lab, and hope that one of them works in humans.
Consider this: 68% of all sequence data in the NCBI Sequence Read Archive comes from just five species 2. More than 99.999% of all microbes remain undiscovered 3. We have been training our models, designing our drugs, and building our understanding of life from a dataset that is, statistically speaking, almost empty.
In January 2026, Basecamp Research released EDEN (Environmentally-Derived Evolutionary Network), "a 28 billion parameter model trained on 9.7 trillion nucleotide tokens from BaseData" 4. It is not just another foundation model. It is a bet that the key to designing therapeutics lies in the raw evolutionary record encoded across millions of species we have never studied -- and that you can't train a model on data you don't have. The early results are striking: AI-designed enzymes that insert genes at specific sites in the human genome, antimicrobial peptides that kill drug-resistant bacteria at a 97% hit rate, and synthetic microbiomes generated at gigabase scale.
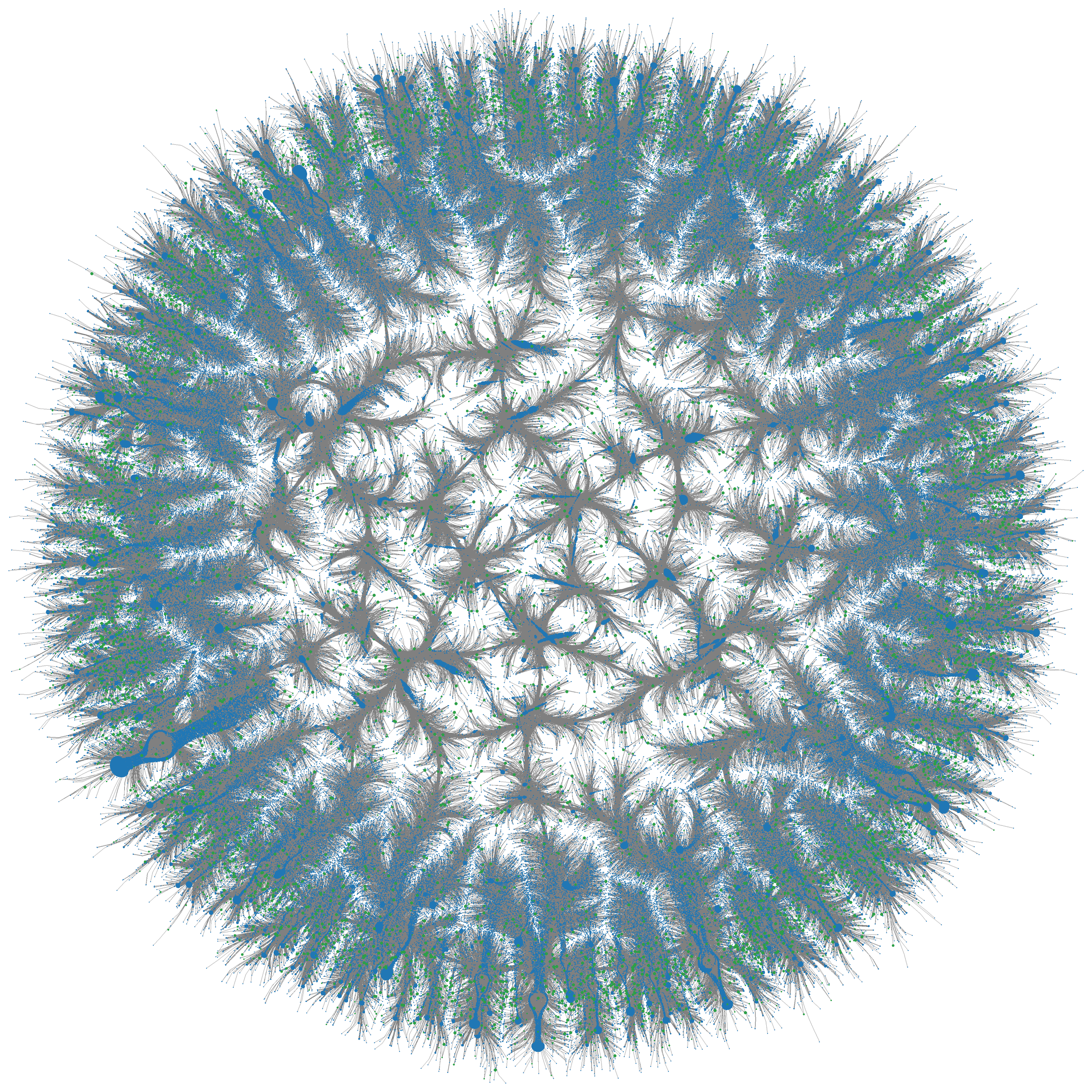 Figure 1: Multi-million-node graph of Large Serine Recombinases (LSRs) from BaseData, clustered at 80% sequence identity. Each node represents an LSR connected to its paired attachment sites -- the raw material EDEN learns from. Source: Munsamy et al., bioRxiv 2026
Figure 1: Multi-million-node graph of Large Serine Recombinases (LSRs) from BaseData, clustered at 80% sequence identity. Each node represents an LSR connected to its paired attachment sites -- the raw material EDEN learns from. Source: Munsamy et al., bioRxiv 2026
What EDEN Is
At its core, EDEN is an autoregressive language model for DNA. If you've worked with GPT-style architectures, the bones are familiar: "EDEN uses a Llama3-style architecture, scaling from 100 million to 28 billion parameters" 4, trained with "a next-token prediction objective and a context length of 8192 tokens" 4. The tokens are nucleotides (the A, C, G, T building blocks of DNA), and the "language" is the functional grammar of genomes.
The model family spans four scales. The team "trained three pairs of EDEN models with 100M, 1B, and 7B parameters on a randomly sampled subset of contigs covering 350 billion nucleotides" 4 to establish scaling laws before committing to the full 28B run. That final model was "trained on up to 1.95x10^24 FLOPs" 4 across "1,008 NVIDIA Hopper GPUs" 5, accelerated with NVIDIA BioNeMo libraries. In compute terms, that places it "among the most computationally intensive biological models ever reported," comparable in scale to GPT-4 class models 5.
 Figure 2: UMAP projection comparing metagenomic assemblies from BaseData (green) and OpenGenome2 (blue), illustrating the expanded sequence space that EDEN's proprietary data covers. Source: Munsamy et al., bioRxiv 2026
Figure 2: UMAP projection comparing metagenomic assemblies from BaseData (green) and OpenGenome2 (blue), illustrating the expanded sequence space that EDEN's proprietary data covers. Source: Munsamy et al., bioRxiv 2026
One detail that caught my attention: there is "no human, lab or clinical data in the pre-training dataset" 4. The model learns biology purely from evolutionary data -- environmental metagenomes, phage sequences, mobile genetic elements. The therapeutic capabilities emerge only at fine-tuning time, when the model's learned understanding of biological grammar gets pointed at specific design tasks. Everything it knows about making therapeutics, it learned from evolution.
The Data That Makes It Different
The single most important thing about EDEN is not its architecture. It is BaseData.
More than 360 biological foundation models have been published to date 2. Most train on public databases. But those databases have a severe problem: "68% of all sequence data in the SRA database coming from just 5 species, this is one of the most severe class imbalance problems ever encountered in AI" 2. When your model has seen the same handful of genomes millions of times and barely touched the rest of life's diversity, it develops blind spots.
Basecamp Research set out to fix this by building BaseData, which they describe as "the largest and fastest-growing biological sequence database ever built, and the first purpose-built for training foundation models" 2. The dataset contained "9.8 billion novel genes, representing more than a 10-fold expansion in known protein diversity" 2 and "more than 1 million species not represented in other genomic databases" 2. It was assembled through physical sample collection -- "partnerships in 18 countries with biodiversity hotspots" 3, with data "collected over five years from over 150 locations across 28 countries and five continents" 5. The database is "capable of growing at over 2 billion genes per month" 4.
 Figure 3: UMAP projection of metagenomic assemblies from BaseData, showing the diversity of genomic data used to train EDEN. Each cluster represents a distinct ecological niche. Source: Munsamy et al., bioRxiv 2026
Figure 3: UMAP projection of metagenomic assemblies from BaseData, showing the diversity of genomic data used to train EDEN. Each cluster represents a distinct ecological niche. Source: Munsamy et al., bioRxiv 2026
Why does this matter for model quality? Two concrete reasons.
First, data quality. BaseData assemblies have a "median contig length of 18.6 kb" compared to "4.0kb for OG2 (metagenomic)" 4, and a "median of 4.9 ORFs per assembly in OG2 (metagenomic) and 20.2 in BaseData" 4. Longer contigs with more open reading frames mean the model sees genes in their natural neighborhood -- surrounded by regulatory elements, neighboring genes, and operon structures -- rather than as isolated fragments. This is the difference between reading a sentence in isolation versus reading it in a paragraph.
Second, scaling efficiency. The team showed that "perplexity decreases more rapidly with compute on BaseData" 4 compared to public data, and "the 7B model trained on BaseData achieves lower test perplexity than its OG2 counterpart" 4. Better data yields better models per FLOP. The data is doing real work here, not just the model size.
The underlying infrastructure is organized around BaseGraph, a knowledge graph "containing over 5.5B relationships with a genomic context exceeding 70 kilobases per protein" 3. Basecamp doesn't just collect sequences. They pair every protein and genome sequence with environmental and chemical data, creating biological context that most databases lack. As CTO Phil Lorenz put it: "Life works as a network, not as a list" 6. And crucially, BaseGraph has "30x more of these LSRs" (large serine recombinases) than public databases 6, including "gene-editing systems used in therapeutic applications that have 0% sequence similarity to anything in public databases" 6.
 Figure 4: BaseData biodiversity catalog -- systematically cataloging genomic dark matter from environmental metagenomes across five continents. Source: Vince et al., Basecamp Research 2025
Figure 4: BaseData biodiversity catalog -- systematically cataloging genomic dark matter from environmental metagenomes across five continents. Source: Vince et al., Basecamp Research 2025
From Sequence to Therapeutic
The EDEN team "challenged a single architecture to design biological novelty across three distinct therapeutic modalities, disease areas and biological scales" 4. This breadth matters. Most biological AI models specialize in one task. EDEN tries to be a general-purpose therapeutic design engine.
Gene Insertion: Programming Where DNA Goes
"Programmable Gene Insertion (PGI), the ability to efficiently insert large pieces of DNA into specific genomic locations, is a significant challenge for the gene editing field" 4. CRISPR gets the headlines, but it works by cutting DNA -- creating double-strand breaks that the cell must repair. Large Serine Recombinases (LSRs) offer an alternative. They "can efficiently integrate large pieces of DNA (>30 kb)" and "are not reliant on DNA damage or host DNA repair pathways, meaning they are efficient in both dividing and quiescent cells" 4. The problem is that "only a few LSRs, such as Bxb1 and PhiC31, have been characterized to date, with limited efficiency as tools for DNA integration in human cells" 7.
EDEN flips this. Instead of searching nature for an LSR that happens to target the site you want, you prompt the model with 30 nucleotides of your target site and it designs an enzyme to match. The model was fine-tuned to "generate de novo large serine recombinase proteins when prompted with only the desired genomic target site" 4.
The results tell the story in stages. First, the hit rate: "EDEN achieves an overall functional hit rate of 63.2% across diverse DNA prompts when prompted on only 30bp of DNA from outside the training data" 4. Then, the coverage: EDEN "generated multiple active recombinases for all tested disease-associated genomic loci (ATM, DMD, F9, FANCC, GALC, IDS, P4HA1, PHEX, RYR2, USH2A) and 4 potential safe harbor sites in the human genome" 4. That is 100% of tested disease targets. The novelty: "several high-performing candidates shared as little as 52% sequence identity with the parental protein, indicating that EDEN is learning biological 'grammar'" 4 -- not copying nature, but composing novel solutions. And "top tier variants exhibit biochemical activity on par with any natural recombinases screened to date" 4.
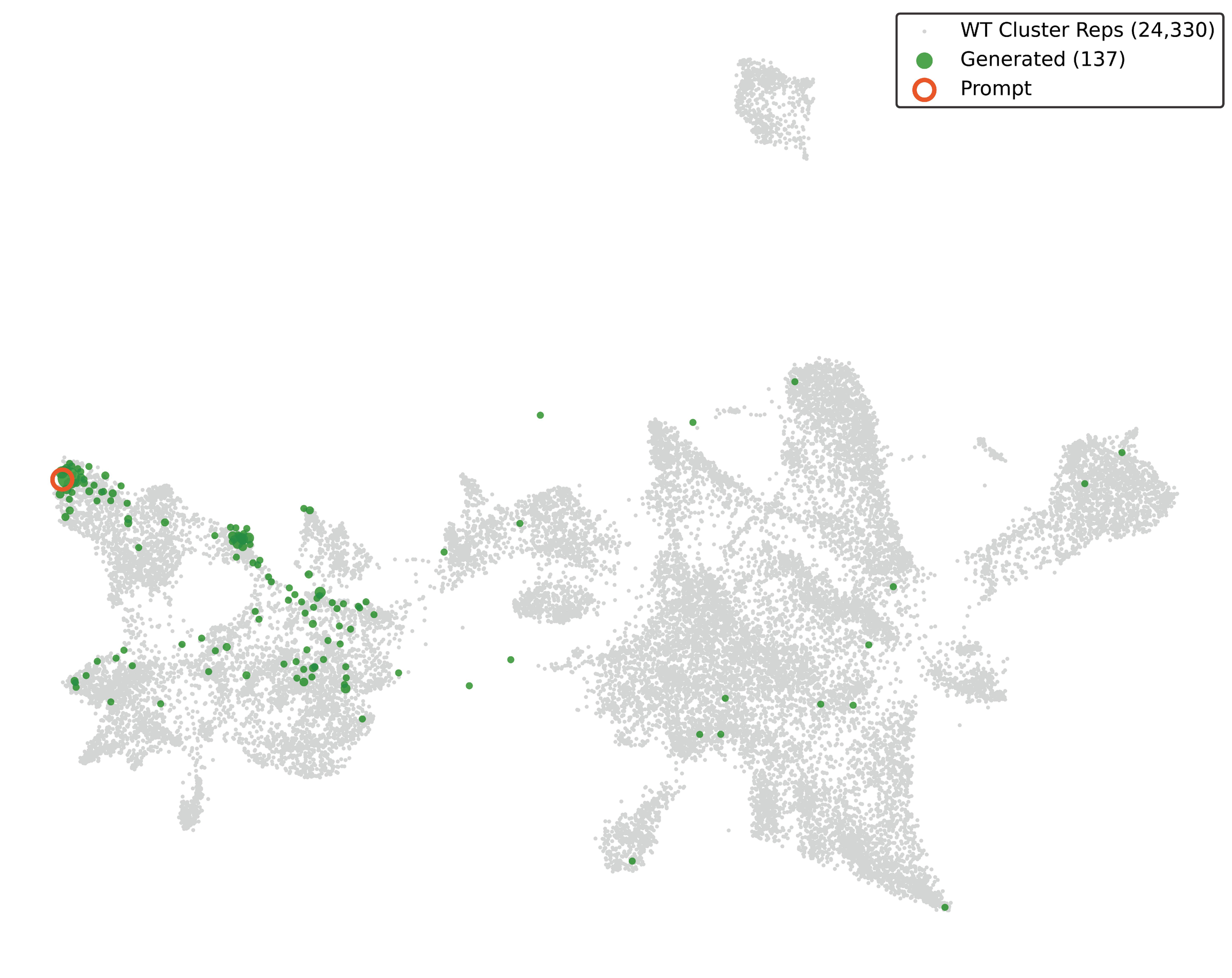 Figure 5: UMAP showing EDEN-generated LSRs (green) distributed across the wild-type sequence space (grey, 24,330 cluster representatives). The model generates novel enzymes that explore diverse regions of LSR sequence space rather than clustering near the training prompt. Source: Munsamy et al., bioRxiv 2026
Figure 5: UMAP showing EDEN-generated LSRs (green) distributed across the wild-type sequence space (grey, 24,330 cluster representatives). The model generates novel enzymes that explore diverse regions of LSR sequence space rather than clustering near the training prompt. Source: Munsamy et al., bioRxiv 2026
The clinical relevance comes from CAR-T results. "50% of EDEN-generated LSRs were active in human cells, achieving therapeutically relevant levels of CAR insertion in primary human T cells" 4. Those engineered T cells showed "over 90 percent cancer cell clearance" in laboratory assays 8. This is the pipeline: take a patient's T cells, use an EDEN-designed recombinase to insert a cancer-fighting gene, put the cells back.
EDEN also demonstrated a second gene-insertion pathway: "EDEN can generate active bridge recombinases when prompted on the associated guide RNA alone, with sequence identities to training and public data as low as 65%" 4. Bridge recombinases are a distinct class of RNA-guided DNA recombinases 9 that offer yet another mechanism for programmable genome editing, broadening the toolkit beyond LSRs.
Antimicrobial Peptides: Fighting Superbugs
"The discovery of novel antimicrobial peptides (AMPs) against clinical superbugs is urgently needed to address the ongoing antibiotic resistance crisis" 10. AMPs are attractive because of "their broad-spectrum activity, rapid bactericidal mechanisms and reduced likelihood of inducing resistance compared with conventional antibiotics" 10.
EDEN's results: "32 of the 33 EDEN designed peptides (97%) were functional" 4, "with top candidates achieving single-digit micromolar potency against critical-priority multidrug-resistant pathogens" 4. This "marks the first instance a DNA foundation model has been used directly for peptide and antibiotics design with proven potency in ground-truth experiments against targets of interest" 11.
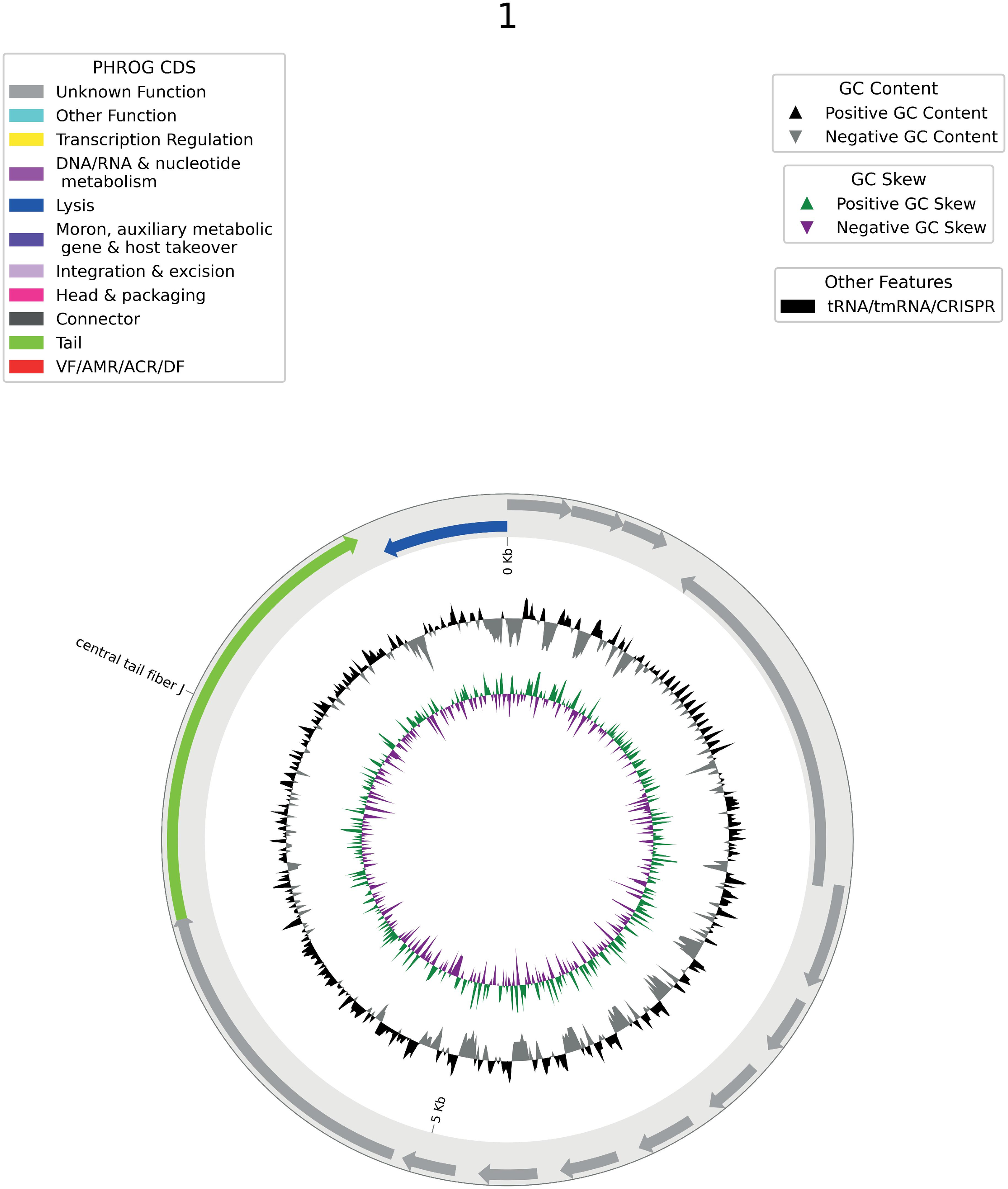 Figure 6: A representative EDEN-generated synthetic prophage genome, showing gene annotations by functional category (PHROG CDS), GC content, and GC skew patterns consistent with real bacteriophage biology. Source: Munsamy et al., bioRxiv 2026
Figure 6: A representative EDEN-generated synthetic prophage genome, showing gene annotations by functional category (PHROG CDS), GC content, and GC skew patterns consistent with real bacteriophage biology. Source: Munsamy et al., bioRxiv 2026
To put that 97% in context: a latent diffusion model pipeline yielded 25 active peptides out of 40 synthesized (62.5%) 12. A machine learning scan of the global microbiome identified 863,498 candidate AMPs, of which 79 of 100 tested peptides were active 13. EDEN's near-saturation hit rate from a DNA foundation model -- not a specialized protein model -- suggests the model has learned something real about antimicrobial function from environmental data. Which makes sense, given that environmental microbiomes have been waging chemical warfare for billions of years.
Synthetic Microbiomes: Designing Ecosystems
The most ambitious application operates at the community scale. EDEN was used to "design a gigabase-scale microbiome with over 94,000 synthetic metagenomic assemblies, including prophage genomes and correct cross-species metabolic pathway completions" 4.
 Figure 7: EDEN generates gigabase-scale synthetic microbiomes that recapitulate the taxonomic and functional diversity of natural digestive system communities. Source: Munsamy et al., bioRxiv 2026
Figure 7: EDEN generates gigabase-scale synthetic microbiomes that recapitulate the taxonomic and functional diversity of natural digestive system communities. Source: Munsamy et al., bioRxiv 2026
The generated microbiome covers "9,067 species with a biome-specific taxonomic accuracy of 99%" 4, and "over 1,500 of the generated species were outside the fine-tuning dataset while retaining the correct microecological properties and biome association" 4. The model generated species it had never seen during fine-tuning, and those species still had the right ecological properties for their assigned biome.
"By generating a microbiome that coheres at the gigabase scale, we show that EDEN captures statistical regularities at the metagenome level" 4. The model has learned something about how microbial communities organize themselves -- which species coexist, how metabolic pathways distribute across community members, how phage populations integrate with their hosts. This is not just generating individual genes. It is generating ecology.
How It Compares
EDEN enters a field with "more than 360 biological foundation models" published to date 2. Rather than catalog each competitor, it's worth asking what bets they represent -- because each major model makes a different wager about what matters most for biological AI.
The architectural novelty bet: Evo 2. Arc Institute's "40-billion parameter genomic foundation model" 14 bets that architecture matters. Evo 2 uses the StripedHyena architecture, "which scales sub-quadratically with context length" 14, enabling a 1M-token context window versus EDEN's 8K. Trained on "9.3 trillion tokens of DNA assembled from 128,000 complete genomes" 14, Evo 2 is fully open-source and excels at long-range genomic understanding. The trade-off: it trains on public data, and hasn't demonstrated direct therapeutic design with wet-lab validation.
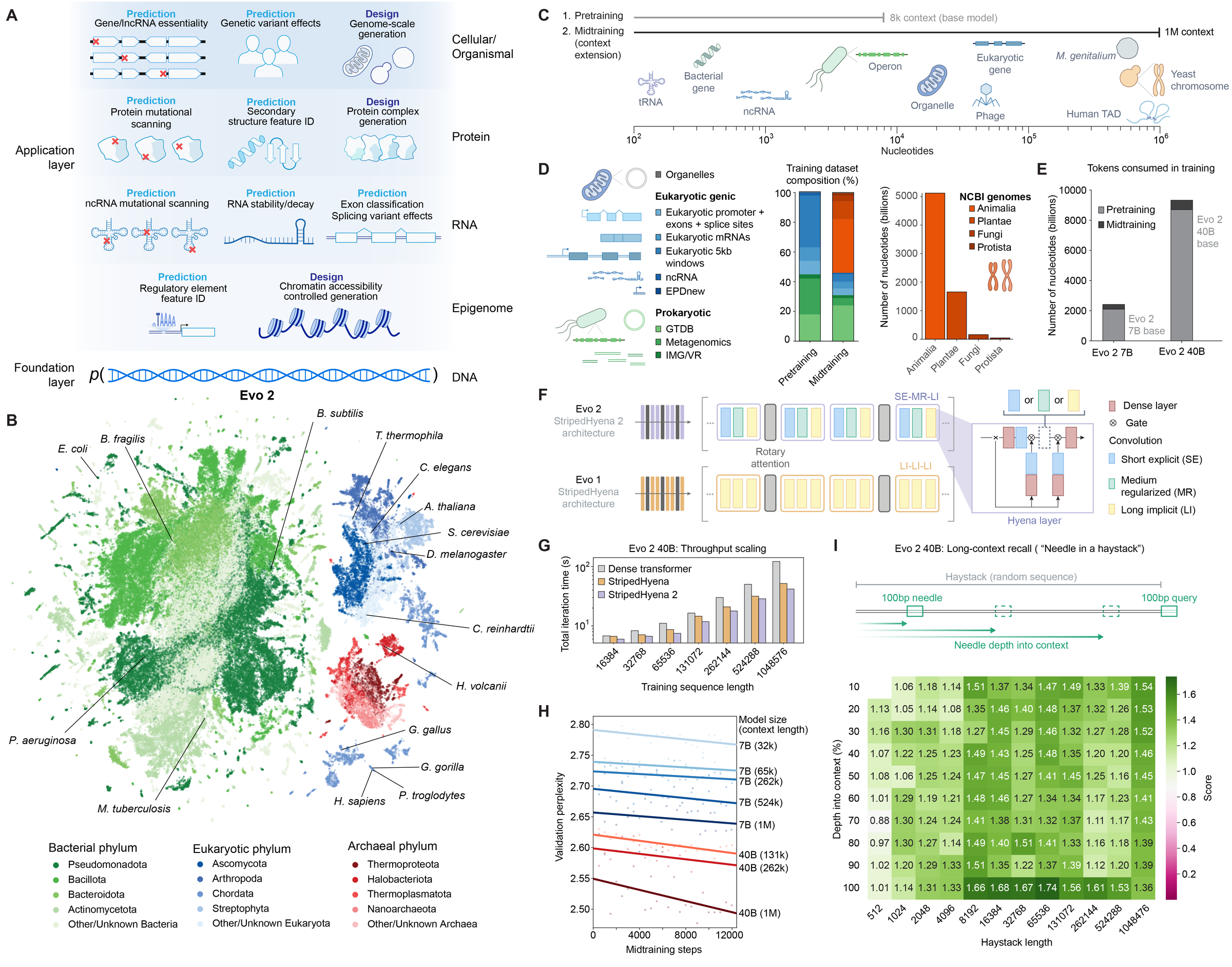 Figure 8: Evo 2 architecture from Arc Institute -- a 40B-parameter genomic foundation model representing the state of the art in open-source DNA language modeling. Source: Brixi et al., bioRxiv 2025
Figure 8: Evo 2 architecture from Arc Institute -- a 40B-parameter genomic foundation model representing the state of the art in open-source DNA language modeling. Source: Brixi et al., bioRxiv 2025
The multimodal structure bet: ESM3. EvolutionaryScale's 98B-parameter model is "a frontier multimodal generative model that reasons over the sequence, structure, and function of proteins" 15. ESM3 natively understands protein structure -- its landmark result was generating esmGFP, a novel fluorescent protein. But ESM3 operates at the protein level, not the nucleotide level. It cannot design genomic features like operons, regulatory elements, or DNA-level editing tools.
The generation-first bet: ProGen and EvoDiff. Salesforce's ProGen demonstrated that "artificial proteins generated by ProGen are as active and thermostable as natural lysozymes" 16 using a 1.2B parameter model. Microsoft's EvoDiff takes a different approach entirely: "a general-purpose diffusion framework for generating protein sequences and structures" 17 where "unlike autoregressive models, EvoDiff can generate proteins with desired structural or functional properties through various conditioning strategies without task-specific fine-tuning" 17. Both are protein-only.
EDEN's bet is on data diversity. Not raw parameter count -- Evo 2 is larger, ESM3 much larger. Not architectural innovation. The wager is that proprietary environmental metagenomics, with 4x more genomic context per assembly than public alternatives, unlocks therapeutic capabilities that no amount of public data will match. And EDEN is the only model in this group to demonstrate functional enzyme design, antimicrobial peptide generation, and microbiome synthesis from a single architecture, all validated in the wet lab.
What This Changes
The EDEN paper makes a strong claim: "the combination of billions of years of evolutionary data with specific therapeutic records offers a clear, scaling-driven path to making therapeutic design a predictable engineering discipline" 4. That word "predictable" is doing heavy lifting. Here is what the evidence supports, and where healthy skepticism is warranted.
What is established. A single foundation model can generate functional therapeutic proteins across multiple modalities when trained on sufficiently diverse evolutionary data. The 63.2% hit rate for novel enzyme design, the 97% for antimicrobial peptides, and the 99% taxonomic accuracy for synthetic microbiomes are all experimentally validated. The insight that "training on more evolutionary data is an important and likely underappreciated part of the path towards unified AI systems" 4 for biology has real evidence behind it now.
The data wall is real and addressable. "Progress of AI in biology is now being limited by the availability of high-quality biological sequence data from nature" 2. Basecamp's approach of physically collecting environmental metagenomics from biodiversity hotspots, rather than scraping the same public databases, produces measurably better models. The scaling laws confirm it. And the team has already "demonstrated insertion at over 10,000 disease-related locations in the human genome" 5.
What is still ahead. Everything clinical. These results are in vitro and in laboratory assays. No EDEN-designed therapeutic has entered clinical trials. The model was trained with "no human, lab or clinical data in the pre-training dataset" 4 -- which is both a feature (no data contamination) and a limitation (no learning from clinical outcomes). The gap between "active in a lab assay" and "works as a therapeutic in humans" is measured in years and billions of dollars. The proprietary nature of BaseData means other researchers cannot reproduce or independently build on this work without Basecamp's involvement.
Still, the direction is clear. As co-founder Oliver Vince noted, "the next step is folding in clinical datasets to this powerful underlying model" 18. And as CSO John Finn stated, "We believe we are at the start of a major expansion of what's possible for patients with cancer and genetic disease" 5. Whether EDEN specifically delivers on that vision remains to be seen. But a model that designs functional enzymes from a 30-nucleotide prompt, and those enzymes actually work in human cells, is a meaningful step. Evolution has been running experiments for 4 billion years. We are finally building models big enough to read the results.
References
Footnotes
-
"NVIDIA BioNeMo Explained: Generative AI in Drug Discovery." IntuitionLabs (2025). https://intuitionlabs.ai/articles/nvidia-bionemo-drug-discovery ↩
-
Basecamp Research. "Breaking Through Biology's Data Wall: Expanding Known Tree of Life by 10x." (2025). https://basecamp-research.com/wp-content/uploads/2025/06/Breaking-Through-Biologys-Data-Wall-Expanding-Known-Tree-of-Life-by-10x.pdf ↩ ↩2 ↩3 ↩4 ↩5 ↩6 ↩7 ↩8
-
"Scaling microbial metagenomic datasets." Owlposting (2025). https://www.owlposting.com/p/scaling-microbial-metagenomic-datasets ↩ ↩2 ↩3
-
Munsamy G, Ayres G, Greco C, et al. "Designing AI-programmable therapeutics with the EDEN family of foundation models." bioRxiv (2026). https://www.biorxiv.org/content/10.64898/2026.01.12.699009v1 ↩ ↩2 ↩3 ↩4 ↩5 ↩6 ↩7 ↩8 ↩9 ↩10 ↩11 ↩12 ↩13 ↩14 ↩15 ↩16 ↩17 ↩18 ↩19 ↩20 ↩21 ↩22 ↩23 ↩24 ↩25 ↩26 ↩27 ↩28 ↩29 ↩30
-
"Basecamp Research launches world-first AI models for programmable gene insertion." PR Newswire (January 12, 2026). https://www.prnewswire.co.uk/news-releases/basecamp-research-launches-world-first-ai-models-for-programmable-gene-insertion-302657979.html ↩ ↩2 ↩3 ↩4 ↩5
-
"Basecamp Research." Neo4j Customer Stories (2025). https://neo4j.com/customer-stories/basecamp-research/ ↩ ↩2 ↩3
-
Durrant MG, Perry NT et al. "Systematic discovery of recombinases for efficient integration of large DNA sequences into the human genome." Nature Biotechnology (2023). https://www.nature.com/articles/s41587-022-01494-w ↩
-
"Basecamp Research and NVIDIA Achieve 97% Success in AI Designed Therapies." BioPharma APAC (January 2026). https://biopharmaapac.com/news/97/7438/basecamp-research-and-nvidia-achieve-97-percent-success-in-ai-designed-therapies-with-programmable-gene-insertion.html ↩
-
Durrant MG et al. "Bridge RNAs direct programmable recombination of target and donor DNA." Nature (2024). https://www.nature.com/articles/s41586-024-07552-4 ↩
-
"A generative artificial intelligence approach for the discovery of antimicrobial peptides against multidrug-resistant bacteria." Nature Microbiology (2025). https://www.nature.com/articles/s41564-025-02114-4 ↩ ↩2
-
"UK startup turns planetary biodiversity into AI-generated drug candidates." The Decoder (January 2026). https://the-decoder.com/uk-startup-turns-planetary-biodiversity-into-ai-generated-drug-candidates/ ↩
-
"Artificial intelligence using a latent diffusion model enables the generation of diverse and potent antimicrobial peptides." Science Advances (2024). https://pubmed.ncbi.nlm.nih.gov/39561365/ ↩
-
de la Fuente-Nunez C et al. "Discovery of antimicrobial peptides in the global microbiome with machine learning." Cell (2024). https://pubmed.ncbi.nlm.nih.gov/38843834/ ↩
-
Brixi G, Kini M, et al. "Genome modeling and design across all domains of life with Evo 2." bioRxiv (2025). https://www.biorxiv.org/content/10.1101/2025.02.18.638918v1 ↩ ↩2 ↩3
-
EvolutionaryScale. "ESM3: Simulating 500 million years of evolution with a language model." Science (2025). https://www.evolutionaryscale.ai/blog/esm3-release ↩
-
Madani A et al. "Large language models generate functional protein sequences across diverse families." Nature Biotechnology 41, 1099-1106 (2023). https://www.nature.com/articles/s41587-022-01618-2 ↩
-
Alamdari S et al. "Protein generation with evolutionary diffusion: sequence is all you need." bioRxiv (2023). https://www.biorxiv.org/content/10.1101/2023.09.11.556673v1 ↩ ↩2
-
"Basecamp Research Achieves Programmable Gene Insertion with EDEN AI Models." Genetic Engineering & Biotechnology News (January 2026). https://www.genengnews.com/topics/artificial-intelligence/basecamp-research-achieves-programmable-gene-insertion-with-eden-ai-models/ ↩